- Home
- About ACA
- Publications & Resources
- Programs
- Annual Meeting
- Membership
- ACA History Center
- Media Archive
ACA Early Career Scientist Spotlight-24
Questions & Answers With Soyoung BaeWhat is your general area of research? I am a biochemist and a cell and molecular biologist by training. I am most interested in explaining cellular phenomena using structure. The bourgeoning field of structural biology uses both X-ray crystallography, micro ED, and cryo-Electron Microscopy. I am actively using visualization techniques to answer subtle questions about enzyme-substrate and enzyme-inhibitor interactions and their interplay.
When I first started learning X-ray crystallography, I was part of a larger effort in the Tolan and Allen laboratories to solve dozens of structures for use in guiding the design of proprietary inhibitors of a target enzyme. I was astonished by the capacity and clarity of the resulting structures, and the gratification of changing the ligand in ways that were predicted from my X-ray crystallographic structures and finding that potency increased.
Talk about the crystallization challenges? Finding the right conditions and determining what kind of source of electron beam I’d need to use (e.g. neutron diffraction vs. synchrotron vs. classical X-ray) is the hardest part of obtaining a crystallization dataset.
The first crystal structure was of an enzyme that is a target for drug development. This was summer(AUG) of 2021, and Clarke Gasper, a former PhD student in Tolan laboratory helped me with crystallization conditions and data collection for the particular co-complexed protein structure. Data was collected from the local Bruker source at the time, with Proteum 3 software. Clarke helped me with the phasing and molecular replacement for the first time so that I could finish the structure. I was astonished to see all the residues I had learned about in biochemistry, and all the structural elements. Since that first structure, I have now solved nearly a hundred structures of KHK enzymes from different species, isozymes, and bound to various ligands. Most of the work has been proprietary, so it will take some time to get published. In the panels below are a handful of the beautiful crystals I have obtained.
What are some of the other projects going on in your group? Who else is in your group? As mentioned, I am part of a large group of scientists, who are all interested in structural biology. The group comprises members of the Tolan and Allen labs. Dr. Tolan, who is a member of the Biology Department, and Dr. Allen, who is a member of the Chemistry Department, have been collaborating for over 25 years and have jointly trained several graduate students and post-docs, published together extensively, and have joint grant awards. In conjunction of Dr. Barbara Imperiali at MIT, there is a major effort towards understanding substrate specificity, structural characteristics, and structure/function relationships of membrane-bound phosphoglycosyl transferases (PGT) from bacterial glycoconjugate biosynthetic pathways. There are also several projects that aim at investigation of protein-protein interactions, as well as projects similar to mine in developing small molecule inhibitors against the potent neurotoxin produced by the soil-dwelling bacterium Clostridium botulinum. In the Tolan lab, there is an effort to look at protein-protein complexes and their interactions among various transcription factors involved in the signal transduction pathway of human KHK-A in its role as a protein kinase. Also, the structure of aldolase A complexed with actin filaments in attempts to bring about an understanding of aldolase in a moonlighting role.
What is your favorite spot within your university? In my lab, I feel the most safe and capable of investigating, utilizing basic science and performing scientific methods. What is something that the last year has taught you? Delayed gratification may be something I am appreciating, as this year I am beginning to see how what seemed complicated and complex at work seems to be viewed rather clearer over time. I think I might be beginning to see what is rather auxiliary in my work, what should be the main story to tell, and what warrants further investigation. How did you create your poster? I created my ACA poster (poster #356) based on my completed Master’s work in the MCBB program at Boston University. I had been working on the differences among ketohexokinase enzymes from different species and the two isozymes. This entailed a classic ligand-fit study. When I did my first structures of ketohexokinase-A I was surprised by my observation of definite conformational changes, which at that point had not been reported. This observation was the basis of my master’s thesis, which I expanded on while I was writing and completing that degree. This ligand-fit study is nearing completion and along with more mechanistic analysis of the mode of binding and catalysis for this protein I hope to publish the entire story with the next year or so. Is there any software program that you think other researchers should know about? During my time delving into X-ray crystallography, I was also looking into other programs and algorithms that might be useful for me in structural biology. I had done some cell culture experiments, including some transcriptomics, which I recently reported at the joint American Society for Cell Biology|European Molecular Biology Organization (ASCB|EMBO). From the ASCB|EMBO meeting, I got to learn an algorithm written to conduct a similarity network not only primarily based on sequence congruency but also by prioritizing structural homology. That program is Protein Cartography by ARCADIA. I heard a couple of other companies are pursuing developing such interactive structure based maps. Also, a web server, the surface entropy reduction prediction server (SERp server) incorporates a conformational entropy profile, a likelihood of secondary structure, and sequence conservation and outputs desirable mutation sites in aim to minimize the entropic price of conformational change, thus most stabilized, suited thermodynamically, therefore, a protein to crystallize- in theory). A recent hot topic in the field is the generative AI driven prediction software for cryoEM, CryoDragon. While the program itself may predict sets of potential dynamics of a macromolecule, it self-corrects its own prone error to come up with the next sets of most plausible repertoire. What inspires your research? Seeing the truth. The fact that data doesn’t lie and it reflects the diligence that goes into the well-implemented, controlled experiments when evidence is captured. What are some of your favorite non-science activities? Music and art. I really enjoy learning about art and architectural histories to appreciate how humanity was able to build from scratch, by rationality and knowledge. At spare time, I enjoy getting violin lessons and being coached in chamber or orchestral music so that I can improve my music, which feeds my esthetics and desire to melt and implement art into my science. I also enjoy playing tennis, although I am at a beginner’s level. How do you explain crystallography to your non-crystallographer friends and family? It's a way to see proteins, the secret of life. |


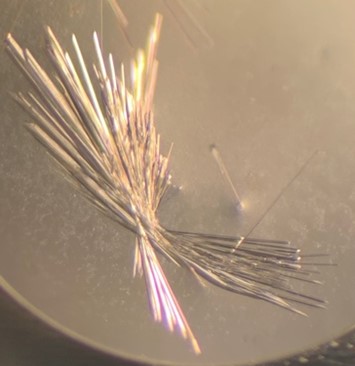
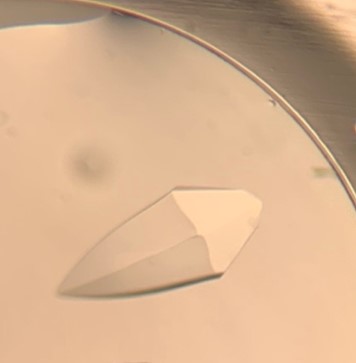
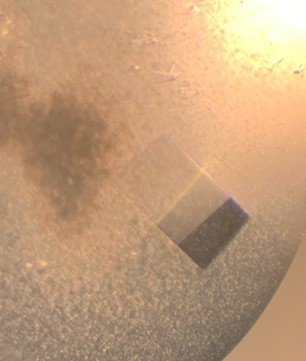

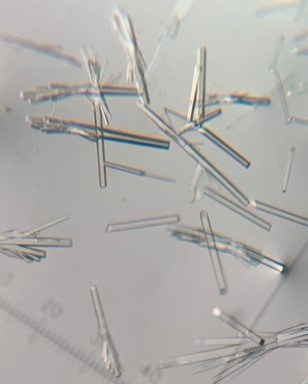

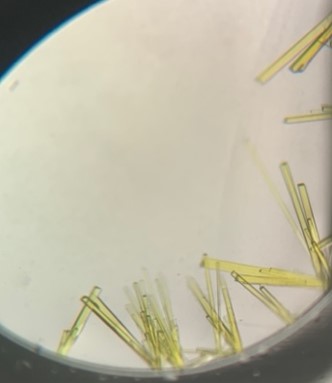
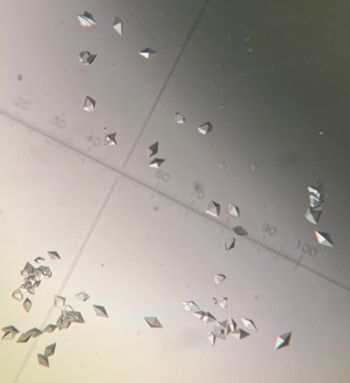
 The instrumentation for structural biology at the BU-Chemistry Instrumentation Center (CIC) includes a new Brüker AXS X8 Proteum-R diffractometer with a Microstar Cu rotating anode X-ray source and a PLATINUM135 charge-coupled device (CCD) area detector. This NIH-sponsored instrument is operated by Dr. Jeff Bacon, an ACA member- someone who has been an integral part of teaching me how to collect better datasets with insight in technical challenges.
The instrumentation for structural biology at the BU-Chemistry Instrumentation Center (CIC) includes a new Brüker AXS X8 Proteum-R diffractometer with a Microstar Cu rotating anode X-ray source and a PLATINUM135 charge-coupled device (CCD) area detector. This NIH-sponsored instrument is operated by Dr. Jeff Bacon, an ACA member- someone who has been an integral part of teaching me how to collect better datasets with insight in technical challenges.


















